Data anlalysis for the project hambiTippingPoints
Manuscript:
Background
The effects of antimicrobial therapies on the human gut microbiome is a critical issue for global health. A typical consequence of antimicrobial therapy is microbiome dysbiosis - the alteration of microbiome structure from a “healthy” state. A human microbiome in dysbiosis can provide growth opportunities for pathogenic and antimicrobial-resistant bacterial strains, which are global threats to human health. Alternatively, an ecologically resilient microbiome resists antimicrobial disturbances and pathogenic invasions. Resilience is an ecosystem’s ability to withstand or rebound from disturbance to its original state. Abrupt shifts in the ecosystem state, known as critical transitions, can occur if a disturbance exceeds the ecosystem’s buffering capacity, which pushes the system to an alternative and often drastically different stable state. Critical transitions are well-known phenomena in large-scale ecosystems - e.g., commercial fisheries collapse due to over-harvesting - but largely unexplored in the field of microbiome research. There is a great need to predict when and how microbiomes may transition to compositionally and functionally alternative states in response to pressures ranging from antibiotic treatment to temperature change, flooding, fires, and other stresses resulting from anthropogenic climate change.
One unique property of microbiomes is that species can rapidly evolve in response to stress. This rapid evolution will occur on the same time scale as ecosystem dynamics that determine critical transitions. Thus, any comprehensive predictive model of critical transitions in microbiomes will need to account for internal evolutionary processes affecting species interactions and overall community state. For example could the evolution of antibiotic resistance in response to antibiotic treatment accelerate the transition of a microbiome to an alternative state? Alternatively, might evolution stabilize internal community dynamics buffering against change? These questions are completely unanswered and are the motivation for this project.
Relevant papers:
- Recovery rates reflect distance to a tipping point in a living system
- Generic Indicators for Loss of Resilience Before a Tipping Point Leading to Population Collapse
- Early warning signals of extinction in deteriorating environments
- Isolated cell behavior drives the evolution of antibiotic resistance
- The inoculum effect and band-pass bacterial response to periodic antibiotic treatment
- Eco-evolutionary feedbacks between private and public goods: evidence from toxic algal blooms
Experiment overview
The repo contains data from an experiment with a synthetic community of 23 bacterial species exposed to the model penicillins class antibiotic ampicillin as a stressor. Here we aimed to ask:
- Where are the tipping points leading to population collapse in this system?
- What are the early warning signals / critical slowing down of population collapse?
- What is the role for rapid antibiotic resistance evolution in position ecosystem collapse or resillience?
Experimental evolution
Prior to the main experiment, 23 species were grown and experimentally evolved to have resistance/tolerance to the antibiotic ampicillin. Species were each grown in monoculture in serial transfer (x% volume transfer every x days) in the presence of ampicillin done my Ida-Marija (Same protocol as in APEX experiment - I will copy this over when Johannes determines the protocol).
Experiment setup
Three different communities were constructed using species with different evolutionary histories and ampicillin sensitivity.
- EVO1: In this community sensitive and experimental evolved species were mixed. The idea was to add ampicillin evolved species that are typically rare in this community (< 1% average abundance) to test for the effect of this trait in rare species. Amp-evolved species included HAMBI_0097, HAMBI_0262, HAMBI_1279, HAMBI_1299, HAMBI_1842, HAMBI_1988, HAMBI_2159, HAMBI_2164, HAMBI_2494, HAMBI_2659, HAMBI_2792, HAMBI_3031, and HAMBI_3237.
- EVO2: In this community all 23 species added were ampicillin resistant/tolerant.
- EVO3: In this community all 23 species were in their clonal/ancestral form.
Experiment treatment conditions
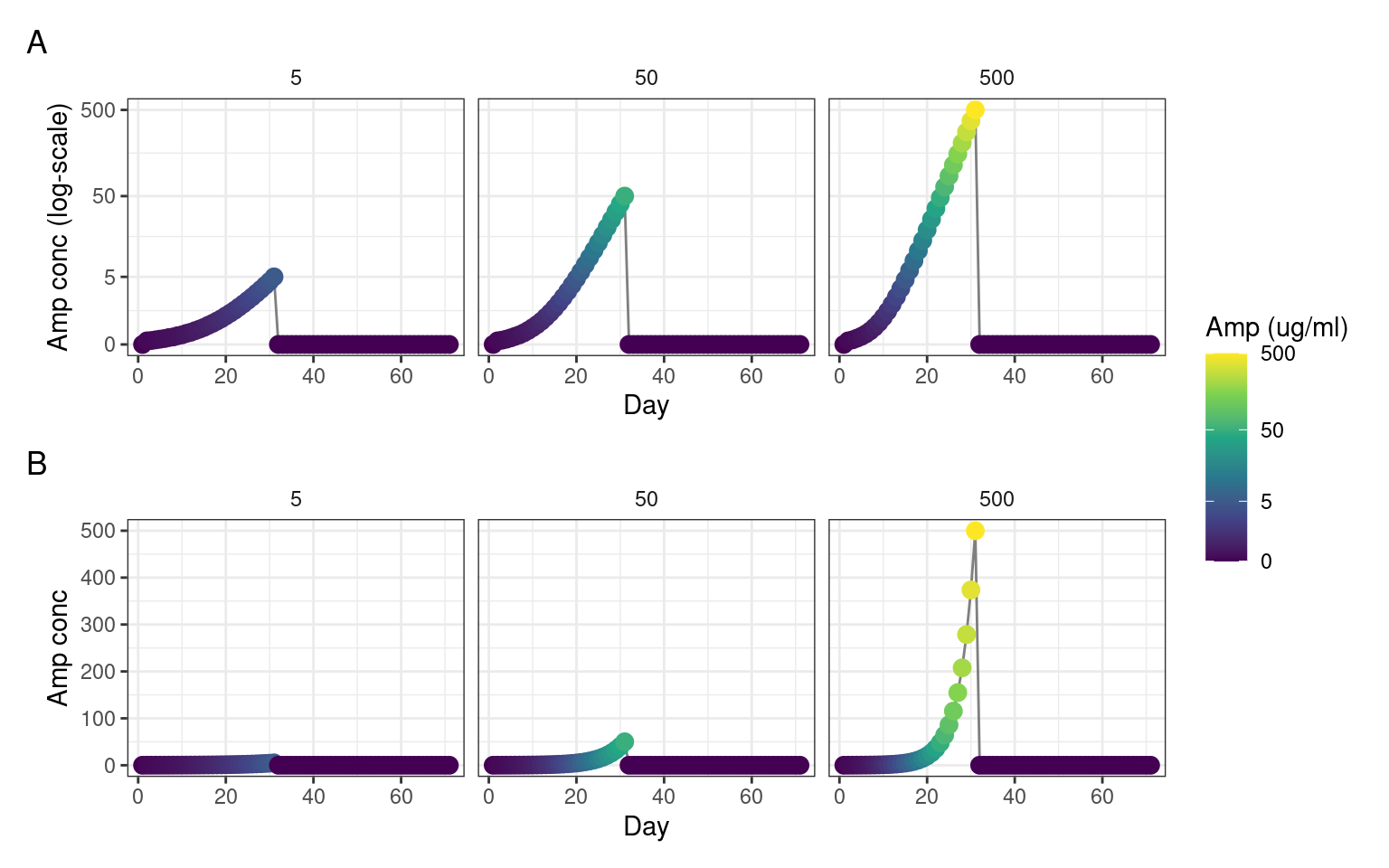
EVO1, EVO2, and EVO3 communities were each grown under three different gradually increasing Ampicillin (AMP) concentrations (endpoints: 5 ug/ml, 50 ug/ml, and 500 ug/ml) + control without AMP. For the first 30 days non-control communities were serially transferred every 24 hours (1/10 dilution, 1 ml total, 96 deep-well plates) to higher AMP concentrations with the AMP endpoints being reached after 30 days - see Figure 1 below. After 30 days, communities were allowed to recover by serially transferring without ampicillin for 41 additional days.
Measurements and data types
- OD600 measured every 24 hours before serial transfer.
- DNA preserved every 24 hours (-20 C) for amplicon sequencing
- Every odd transfer samples were stored 1:1 w/ 85% glycerol in -80C for later clone isolation
Code
Data and code here is provided under GPL3. Feel free to use or remix as you see fit.
Project structure
/Rcontains R scripts/datacontains data that has been processed in some way for later use/_data_rawcontains unprocessed data scraped from compute cluster/figscontains figures generated from R scripts
Availability
The rendered project site is available at https://hambitippingpoints-slhogl-623cfec303619cc856140fccfc094718702f7.utugit.fi/. The website has been produced using Quarto notebooks.
This GitLab repository (https://gitlab.utu.fi/slhogl/hambiTippingPoints) hosts the code and data for this project. The rendered webpage can be fully recreated using the code at https://gitlab.utu.fi/slhogl/hambiTippingPoints.
Reproducibility
The project uses renv to create reproducible environment to execute the code in this project. See here for a brief overview on collaboration and reproduction of the entire project. To get up and running you can do: