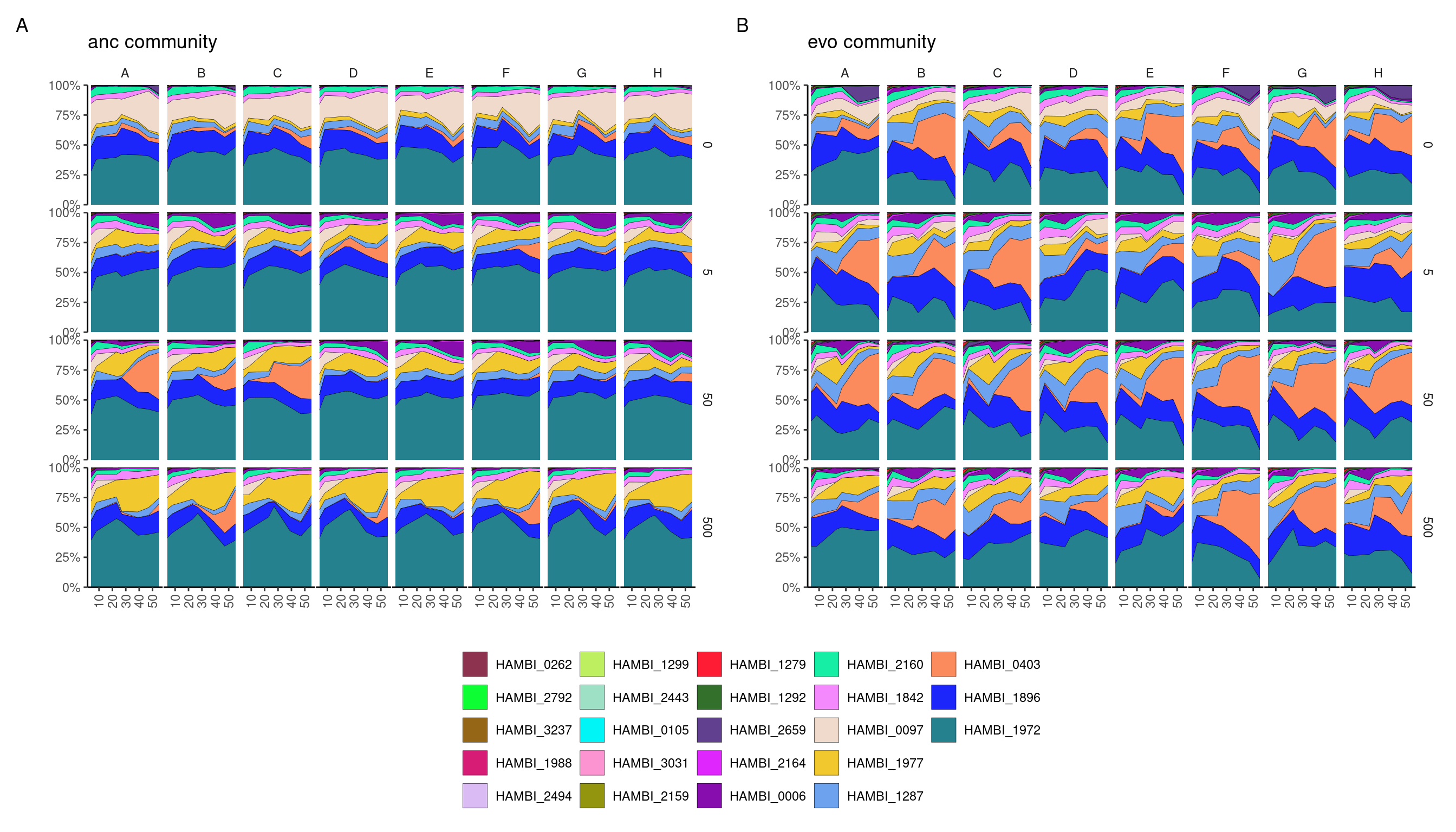
Analysis of community composition
Introduction
Area plots of community composition
Libraries and global variables
Read data
Helps make the area plots look nicer
Plot community compositions from the experiment
phi <- counts %>%
mutate(strainID=factor(strainID, levels=strain.order)) %>%
group_by(evolution) %>%
group_split() %>%
map(myarplot) %>%
wrap_plots(., ncol = 2) +
plot_layout(guides = 'collect') +
plot_annotation(tag_levels = 'A') &
theme(legend.position = 'bottom')Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.